Calculate and optionally plot power curves for different effect sizes and trial counts. This function takes a
Usage
calculate_power_curves(
trials,
effectsize = 1,
candidateset = NULL,
model = NULL,
alpha = 0.05,
gen_args = list(),
eval_function = "eval_design",
eval_args = list(),
random_seed = 123,
iterate_seed = FALSE,
plot_results = TRUE,
auto_scale = TRUE,
x_breaks = NULL,
y_breaks = seq(0, 1, by = 0.1),
ggplot_elements = list()
)Arguments
- trials
A numeric vector indicating the trial(s) used when computing the power curve. If a single value, this will be fixed and only
effectsizewill be varied.- effectsize
Default
1. A numeric vector indicating the effect size(s) used when computing the power curve. If a single value, this will be fixed and onlytrialswill be varied. If using a length-2 effect size witheval_design_mc()(such as a binomial probability interval), the effect size pairs can be input as entries in a list.- candidateset
Default
NULL. The candidate set (seegen_design()documentation for more information). Provided to aid code completion: can also be provided ingen_args.- model
Default
NULL. The model (seegen_design()andeval_design()documentation for more information). Provided to aid code completion: can also be provided ingen_args/eval_args.- alpha
Default
0.05. The allowable Type-I error rate (seeeval_design()documentation for more information). Provided to aid code completion: can also be provided ineval_args.- gen_args
Default
list(). A list of argument/value pairs to specify the design generation parameters forgen_design().- eval_function
Default
"eval_design". A string (or function) specifying the skpr power evaluation function (e.g.eval_design(),eval_design_mc(),eval_design_survival_mc(), oreval_design_custom_mc()).- eval_args
Default
list(). A list of argument/value pairs to specify the design power evaluation parameters foreval_function.- random_seed
Default
123. The random seed used to generate and then evaluate the design. The seed is set right before design generation.- iterate_seed
Default
FALSE. This will iterate the random seed with each new design. Set this toTRUEto add more variability to the design generation process.- plot_results
Default
TRUE. Whether to print out a plot of the power curves in addition to the data frame of results. Requiresggplot2.- auto_scale
Default
TRUE. Whether to automatically scale the y-axis to 0 and 1.- x_breaks
Default
NULL, automaticly generated by ggplot2.- y_breaks
Default
seq(0,1,by=0.1). Y-axis breaks.- ggplot_elements
Default
list(). Extraggplot2elements to customize the plot, passed in as elements in a list.
Examples
if(skpr:::run_documentation()) {
cand_set = expand.grid(brew_temp = c(80, 85, 90),
altitude = c(0, 2000, 4000),
bean_sun = c("low", "partial", "high"))
#Plot power for a linear model with all interactions
calculate_power_curves(trials=seq(10,60,by=5),
candidateset = cand_set,
model = ~.*.,
alpha = 0.05,
effectsize = 1,
eval_function = "eval_design") |>
head(30)
}
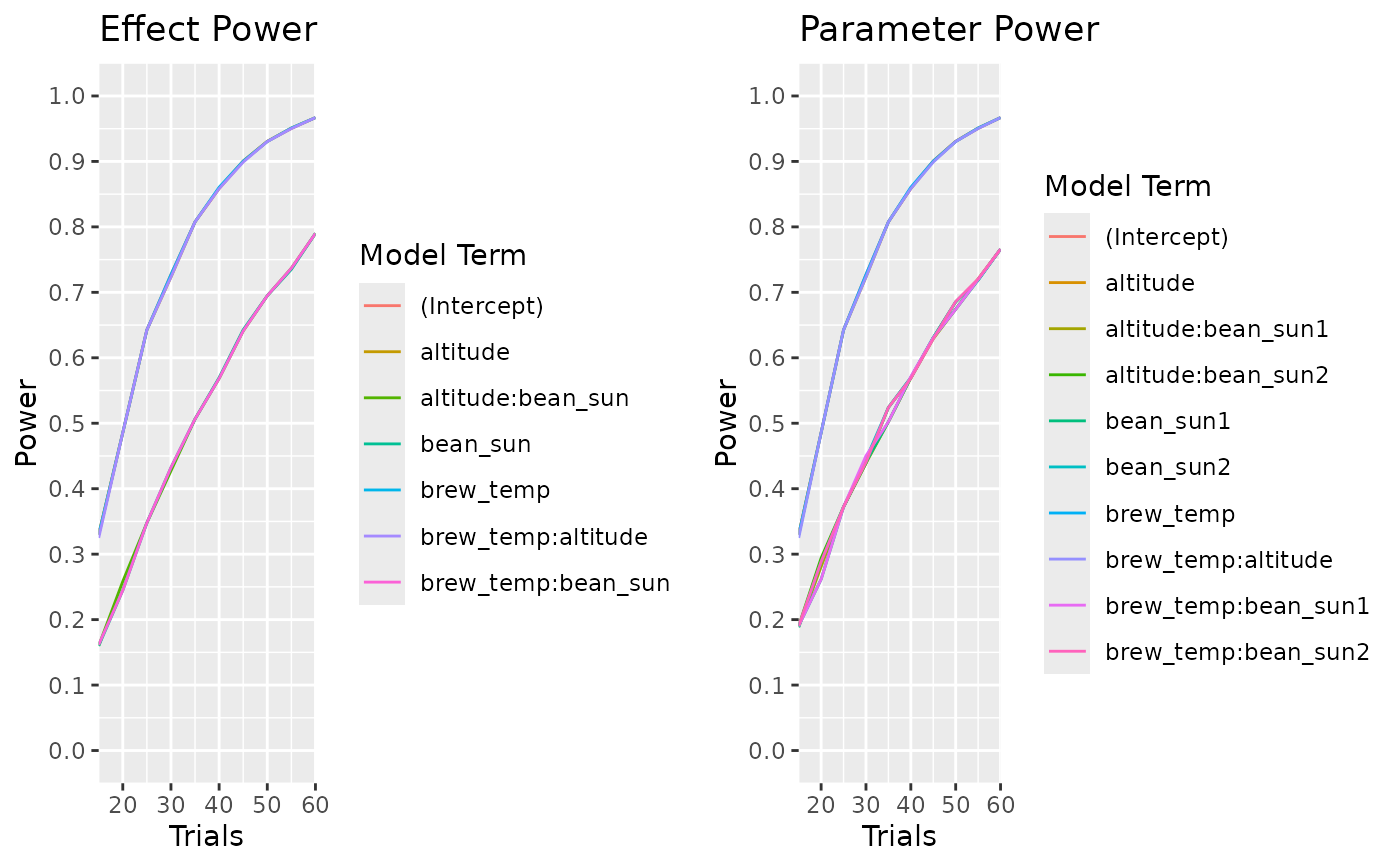 #> parameter type power trials effectsize random_seed
#> 18 (Intercept) effect.power 0.3297088 15 1 123
#> 19 brew_temp effect.power 0.3297088 15 1 123
#> 20 altitude effect.power 0.3297088 15 1 123
#> 21 bean_sun effect.power 0.1599775 15 1 123
#> 22 brew_temp:altitude effect.power 0.3248373 15 1 123
#> 23 brew_temp:bean_sun effect.power 0.1615894 15 1 123
#> 24 altitude:bean_sun effect.power 0.1599775 15 1 123
#> 25 (Intercept) parameter.power 0.3297088 15 1 123
#> 26 brew_temp parameter.power 0.3297088 15 1 123
#> 27 altitude parameter.power 0.3297088 15 1 123
#> 28 bean_sun1 parameter.power 0.1904301 15 1 123
#> 29 bean_sun2 parameter.power 0.1884952 15 1 123
#> 30 brew_temp:altitude parameter.power 0.3248373 15 1 123
#> 31 brew_temp:bean_sun1 parameter.power 0.1904301 15 1 123
#> 32 brew_temp:bean_sun2 parameter.power 0.1904301 15 1 123
#> 33 altitude:bean_sun1 parameter.power 0.1884952 15 1 123
#> 34 altitude:bean_sun2 parameter.power 0.1904301 15 1 123
#> 35 (Intercept) effect.power 0.4856389 20 1 123
#> 36 brew_temp effect.power 0.4856389 20 1 123
#> 37 altitude effect.power 0.4856389 20 1 123
#> 38 bean_sun effect.power 0.2447062 20 1 123
#> 39 brew_temp:altitude effect.power 0.4856389 20 1 123
#> 40 brew_temp:bean_sun effect.power 0.2447062 20 1 123
#> 41 altitude:bean_sun effect.power 0.2583789 20 1 123
#> 42 (Intercept) parameter.power 0.4856389 20 1 123
#> 43 brew_temp parameter.power 0.4856389 20 1 123
#> 44 altitude parameter.power 0.4856389 20 1 123
#> 45 bean_sun1 parameter.power 0.2615680 20 1 123
#> 46 bean_sun2 parameter.power 0.2883561 20 1 123
#> 47 brew_temp:altitude parameter.power 0.4856389 20 1 123
#> Power curve generation captured the following warning/error messages:
#> Function | Type | N | Message
#> Evaluation | Warn | 1 | Message: 'skpr: NA indicates not enough degrees of freedom to estimate power for those terms.'
if(skpr:::run_documentation()) {
#Add multiple effect sizes
calculate_power_curves(trials=seq(10,60,by=1),
candidateset = cand_set,
model = ~.*.,
alpha = 0.05,
effectsize = c(1,2),
eval_function = "eval_design") |>
head(30)
}
#> parameter type power trials effectsize random_seed
#> 18 (Intercept) effect.power 0.3297088 15 1 123
#> 19 brew_temp effect.power 0.3297088 15 1 123
#> 20 altitude effect.power 0.3297088 15 1 123
#> 21 bean_sun effect.power 0.1599775 15 1 123
#> 22 brew_temp:altitude effect.power 0.3248373 15 1 123
#> 23 brew_temp:bean_sun effect.power 0.1615894 15 1 123
#> 24 altitude:bean_sun effect.power 0.1599775 15 1 123
#> 25 (Intercept) parameter.power 0.3297088 15 1 123
#> 26 brew_temp parameter.power 0.3297088 15 1 123
#> 27 altitude parameter.power 0.3297088 15 1 123
#> 28 bean_sun1 parameter.power 0.1904301 15 1 123
#> 29 bean_sun2 parameter.power 0.1884952 15 1 123
#> 30 brew_temp:altitude parameter.power 0.3248373 15 1 123
#> 31 brew_temp:bean_sun1 parameter.power 0.1904301 15 1 123
#> 32 brew_temp:bean_sun2 parameter.power 0.1904301 15 1 123
#> 33 altitude:bean_sun1 parameter.power 0.1884952 15 1 123
#> 34 altitude:bean_sun2 parameter.power 0.1904301 15 1 123
#> 35 (Intercept) effect.power 0.4856389 20 1 123
#> 36 brew_temp effect.power 0.4856389 20 1 123
#> 37 altitude effect.power 0.4856389 20 1 123
#> 38 bean_sun effect.power 0.2447062 20 1 123
#> 39 brew_temp:altitude effect.power 0.4856389 20 1 123
#> 40 brew_temp:bean_sun effect.power 0.2447062 20 1 123
#> 41 altitude:bean_sun effect.power 0.2583789 20 1 123
#> 42 (Intercept) parameter.power 0.4856389 20 1 123
#> 43 brew_temp parameter.power 0.4856389 20 1 123
#> 44 altitude parameter.power 0.4856389 20 1 123
#> 45 bean_sun1 parameter.power 0.2615680 20 1 123
#> 46 bean_sun2 parameter.power 0.2883561 20 1 123
#> 47 brew_temp:altitude parameter.power 0.4856389 20 1 123
#> Power curve generation captured the following warning/error messages:
#> Function | Type | N | Message
#> Evaluation | Warn | 1 | Message: 'skpr: NA indicates not enough degrees of freedom to estimate power for those terms.'
if(skpr:::run_documentation()) {
#Add multiple effect sizes
calculate_power_curves(trials=seq(10,60,by=1),
candidateset = cand_set,
model = ~.*.,
alpha = 0.05,
effectsize = c(1,2),
eval_function = "eval_design") |>
head(30)
}
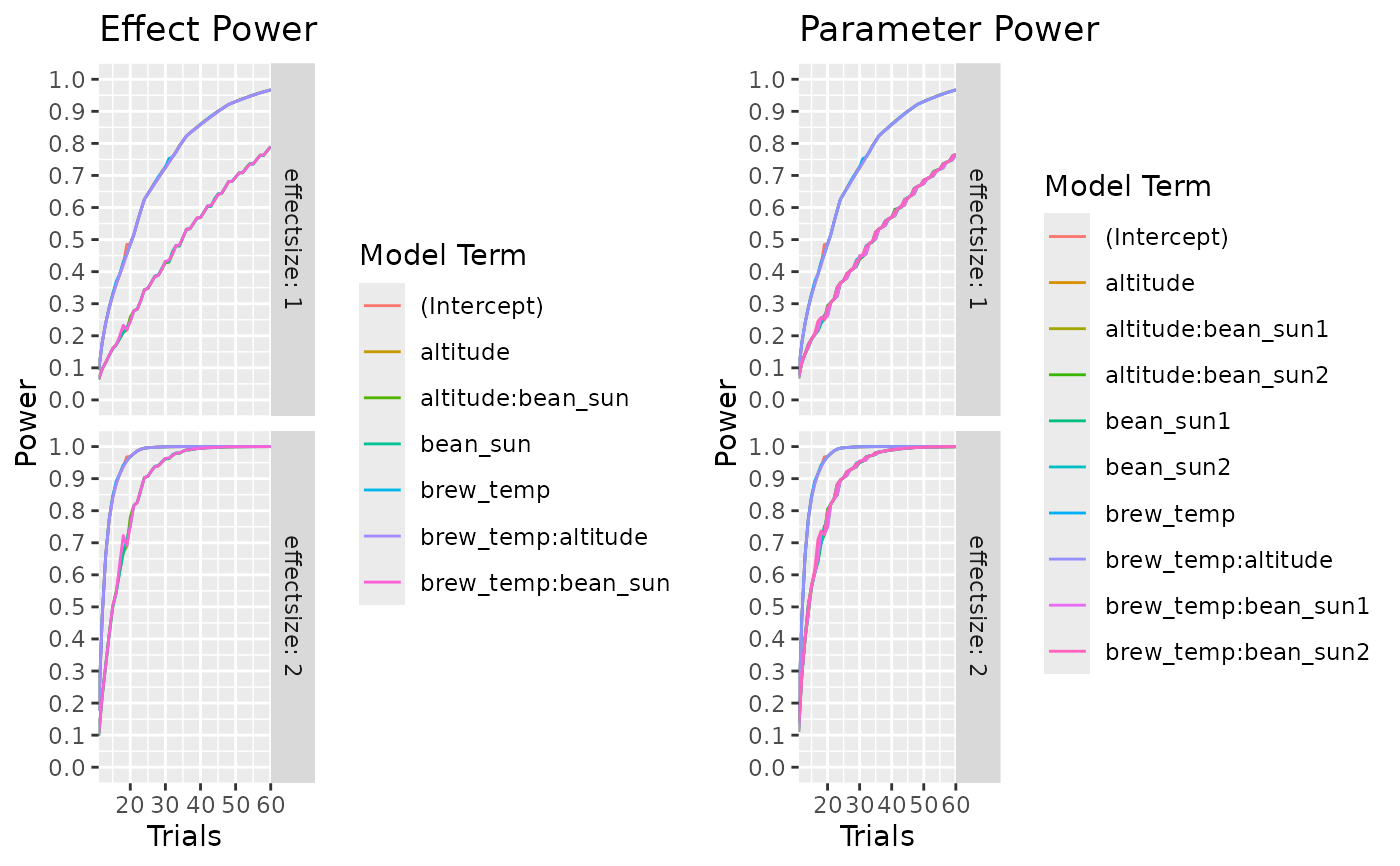 #> parameter type power trials effectsize random_seed
#> 18 (Intercept) effect.power 0.09280916 11 1 123
#> 19 brew_temp effect.power 0.09280916 11 1 123
#> 20 altitude effect.power 0.09280916 11 1 123
#> 21 bean_sun effect.power 0.06447520 11 1 123
#> 22 brew_temp:altitude effect.power 0.09280916 11 1 123
#> 23 brew_temp:bean_sun effect.power 0.06447520 11 1 123
#> 24 altitude:bean_sun effect.power 0.06447520 11 1 123
#> 25 (Intercept) parameter.power 0.09280916 11 1 123
#> 26 brew_temp parameter.power 0.09280916 11 1 123
#> 27 altitude parameter.power 0.09280916 11 1 123
#> 28 bean_sun1 parameter.power 0.06757424 11 1 123
#> 29 bean_sun2 parameter.power 0.07717846 11 1 123
#> 30 brew_temp:altitude parameter.power 0.09280916 11 1 123
#> 31 brew_temp:bean_sun1 parameter.power 0.06757424 11 1 123
#> 32 brew_temp:bean_sun2 parameter.power 0.07717846 11 1 123
#> 33 altitude:bean_sun1 parameter.power 0.06757424 11 1 123
#> 34 altitude:bean_sun2 parameter.power 0.07717846 11 1 123
#> 35 (Intercept) effect.power 0.17925539 12 1 123
#> 36 brew_temp effect.power 0.17925539 12 1 123
#> 37 altitude effect.power 0.17925539 12 1 123
#> 38 bean_sun effect.power 0.09633205 12 1 123
#> 39 brew_temp:altitude effect.power 0.17925539 12 1 123
#> 40 brew_temp:bean_sun effect.power 0.09633205 12 1 123
#> 41 altitude:bean_sun effect.power 0.09633205 12 1 123
#> 42 (Intercept) parameter.power 0.17925539 12 1 123
#> 43 brew_temp parameter.power 0.17925539 12 1 123
#> 44 altitude parameter.power 0.17925539 12 1 123
#> 45 bean_sun1 parameter.power 0.11698959 12 1 123
#> 46 bean_sun2 parameter.power 0.11698959 12 1 123
#> 47 brew_temp:altitude parameter.power 0.17925539 12 1 123
#> Power curve generation captured the following warning/error messages:
#> Function | Type | N | Message
#> Evaluation | Warn | 2 | Message: 'skpr: NA indicates not enough degrees of freedom to estimate power for those terms.'
if(skpr:::run_documentation()) {
#Generate power curve for a binomial model
calculate_power_curves(trials=seq(50,150,by=10),
candidateset = cand_set,
model = ~.,
effectsize = c(0.6,0.9),
eval_function = "eval_design_mc",
eval_args = list(nsim = 100, glmfamily = "binomial")) |>
head(30)
}
#> parameter type power trials effectsize random_seed
#> 18 (Intercept) effect.power 0.09280916 11 1 123
#> 19 brew_temp effect.power 0.09280916 11 1 123
#> 20 altitude effect.power 0.09280916 11 1 123
#> 21 bean_sun effect.power 0.06447520 11 1 123
#> 22 brew_temp:altitude effect.power 0.09280916 11 1 123
#> 23 brew_temp:bean_sun effect.power 0.06447520 11 1 123
#> 24 altitude:bean_sun effect.power 0.06447520 11 1 123
#> 25 (Intercept) parameter.power 0.09280916 11 1 123
#> 26 brew_temp parameter.power 0.09280916 11 1 123
#> 27 altitude parameter.power 0.09280916 11 1 123
#> 28 bean_sun1 parameter.power 0.06757424 11 1 123
#> 29 bean_sun2 parameter.power 0.07717846 11 1 123
#> 30 brew_temp:altitude parameter.power 0.09280916 11 1 123
#> 31 brew_temp:bean_sun1 parameter.power 0.06757424 11 1 123
#> 32 brew_temp:bean_sun2 parameter.power 0.07717846 11 1 123
#> 33 altitude:bean_sun1 parameter.power 0.06757424 11 1 123
#> 34 altitude:bean_sun2 parameter.power 0.07717846 11 1 123
#> 35 (Intercept) effect.power 0.17925539 12 1 123
#> 36 brew_temp effect.power 0.17925539 12 1 123
#> 37 altitude effect.power 0.17925539 12 1 123
#> 38 bean_sun effect.power 0.09633205 12 1 123
#> 39 brew_temp:altitude effect.power 0.17925539 12 1 123
#> 40 brew_temp:bean_sun effect.power 0.09633205 12 1 123
#> 41 altitude:bean_sun effect.power 0.09633205 12 1 123
#> 42 (Intercept) parameter.power 0.17925539 12 1 123
#> 43 brew_temp parameter.power 0.17925539 12 1 123
#> 44 altitude parameter.power 0.17925539 12 1 123
#> 45 bean_sun1 parameter.power 0.11698959 12 1 123
#> 46 bean_sun2 parameter.power 0.11698959 12 1 123
#> 47 brew_temp:altitude parameter.power 0.17925539 12 1 123
#> Power curve generation captured the following warning/error messages:
#> Function | Type | N | Message
#> Evaluation | Warn | 2 | Message: 'skpr: NA indicates not enough degrees of freedom to estimate power for those terms.'
if(skpr:::run_documentation()) {
#Generate power curve for a binomial model
calculate_power_curves(trials=seq(50,150,by=10),
candidateset = cand_set,
model = ~.,
effectsize = c(0.6,0.9),
eval_function = "eval_design_mc",
eval_args = list(nsim = 100, glmfamily = "binomial")) |>
head(30)
}
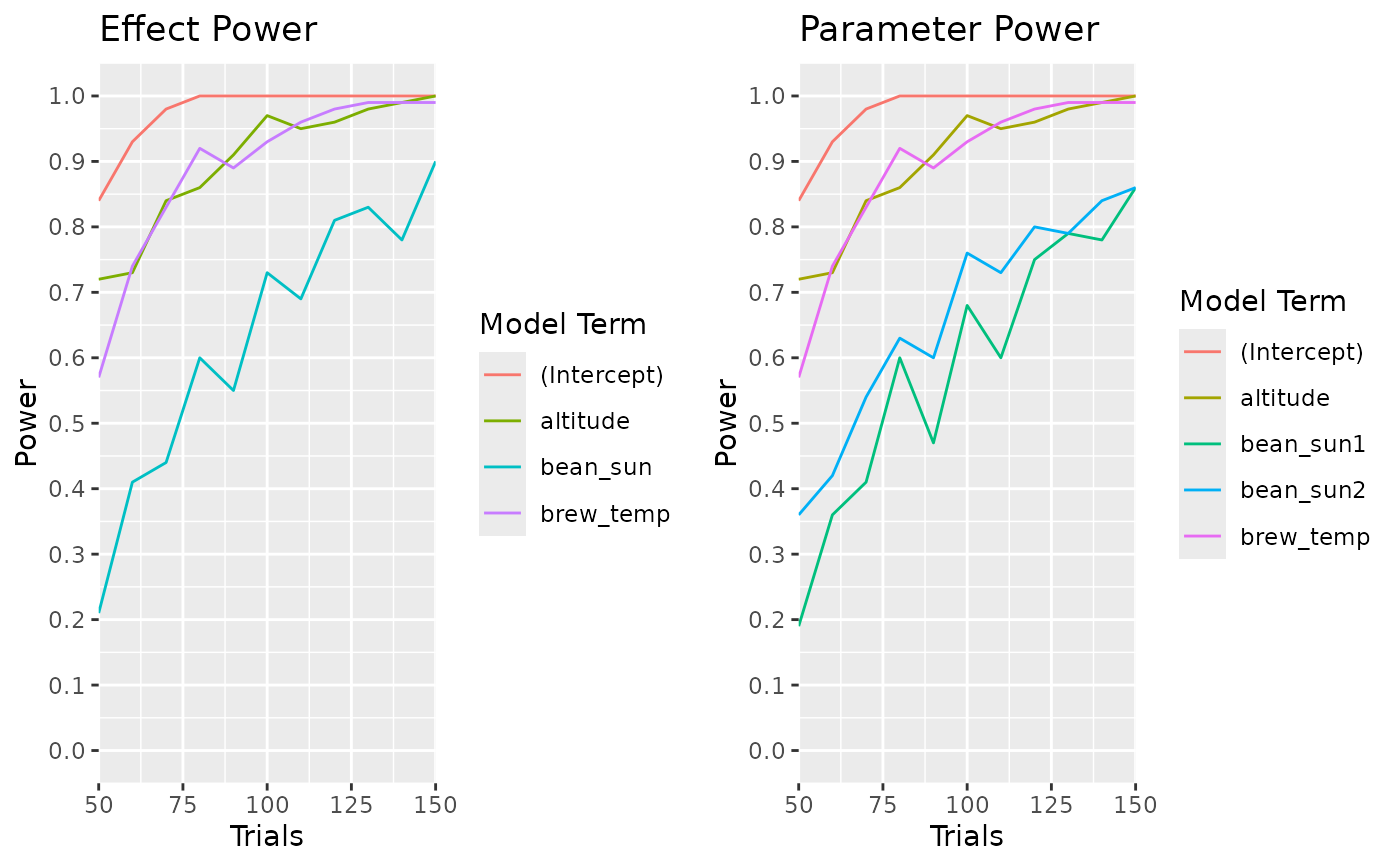 #> parameter type power trials effectsize_low effectsize_high
#> 1 (Intercept) effect.power.mc 0.84 50 0.6 0.9
#> 2 brew_temp effect.power.mc 0.57 50 0.6 0.9
#> 3 altitude effect.power.mc 0.72 50 0.6 0.9
#> 4 bean_sun effect.power.mc 0.21 50 0.6 0.9
#> 5 (Intercept) parameter.power.mc 0.84 50 0.6 0.9
#> 6 brew_temp parameter.power.mc 0.57 50 0.6 0.9
#> 7 altitude parameter.power.mc 0.72 50 0.6 0.9
#> 8 bean_sun1 parameter.power.mc 0.19 50 0.6 0.9
#> 9 bean_sun2 parameter.power.mc 0.36 50 0.6 0.9
#> 10 (Intercept) effect.power.mc 0.93 60 0.6 0.9
#> 11 brew_temp effect.power.mc 0.74 60 0.6 0.9
#> 12 altitude effect.power.mc 0.73 60 0.6 0.9
#> 13 bean_sun effect.power.mc 0.41 60 0.6 0.9
#> 14 (Intercept) parameter.power.mc 0.93 60 0.6 0.9
#> 15 brew_temp parameter.power.mc 0.74 60 0.6 0.9
#> 16 altitude parameter.power.mc 0.73 60 0.6 0.9
#> 17 bean_sun1 parameter.power.mc 0.36 60 0.6 0.9
#> 18 bean_sun2 parameter.power.mc 0.42 60 0.6 0.9
#> 19 (Intercept) effect.power.mc 0.98 70 0.6 0.9
#> 20 brew_temp effect.power.mc 0.83 70 0.6 0.9
#> 21 altitude effect.power.mc 0.84 70 0.6 0.9
#> 22 bean_sun effect.power.mc 0.44 70 0.6 0.9
#> 23 (Intercept) parameter.power.mc 0.98 70 0.6 0.9
#> 24 brew_temp parameter.power.mc 0.83 70 0.6 0.9
#> 25 altitude parameter.power.mc 0.84 70 0.6 0.9
#> 26 bean_sun1 parameter.power.mc 0.41 70 0.6 0.9
#> 27 bean_sun2 parameter.power.mc 0.54 70 0.6 0.9
#> 28 (Intercept) effect.power.mc 1.00 80 0.6 0.9
#> 29 brew_temp effect.power.mc 0.92 80 0.6 0.9
#> 30 altitude effect.power.mc 0.86 80 0.6 0.9
#> random_seed
#> 1 123
#> 2 123
#> 3 123
#> 4 123
#> 5 123
#> 6 123
#> 7 123
#> 8 123
#> 9 123
#> 10 123
#> 11 123
#> 12 123
#> 13 123
#> 14 123
#> 15 123
#> 16 123
#> 17 123
#> 18 123
#> 19 123
#> 20 123
#> 21 123
#> 22 123
#> 23 123
#> 24 123
#> 25 123
#> 26 123
#> 27 123
#> 28 123
#> 29 123
#> 30 123
#> Power curve generation captured the following warning/error messages:
#> Function | Type | N | Message
#> Evaluation | Warn | 1 | Message: 'skpr: Partial or complete separation likely detected in the binomial Monte Carlo simulation. Increase the number of runs in the design or decrease the number of model parameters to improve power.'
if(skpr:::run_documentation()) {
#Generate power curve for a binomial model and multiple effect sizes
calculate_power_curves(trials=seq(50,150,by=10),
candidateset = cand_set,
model = ~.,
effectsize = list(c(0.5,0.9),c(0.6,0.9)),
eval_function = "eval_design_mc",
eval_args = list(nsim = 100, glmfamily = "binomial")) |>
head(30)
}
#> parameter type power trials effectsize_low effectsize_high
#> 1 (Intercept) effect.power.mc 0.84 50 0.6 0.9
#> 2 brew_temp effect.power.mc 0.57 50 0.6 0.9
#> 3 altitude effect.power.mc 0.72 50 0.6 0.9
#> 4 bean_sun effect.power.mc 0.21 50 0.6 0.9
#> 5 (Intercept) parameter.power.mc 0.84 50 0.6 0.9
#> 6 brew_temp parameter.power.mc 0.57 50 0.6 0.9
#> 7 altitude parameter.power.mc 0.72 50 0.6 0.9
#> 8 bean_sun1 parameter.power.mc 0.19 50 0.6 0.9
#> 9 bean_sun2 parameter.power.mc 0.36 50 0.6 0.9
#> 10 (Intercept) effect.power.mc 0.93 60 0.6 0.9
#> 11 brew_temp effect.power.mc 0.74 60 0.6 0.9
#> 12 altitude effect.power.mc 0.73 60 0.6 0.9
#> 13 bean_sun effect.power.mc 0.41 60 0.6 0.9
#> 14 (Intercept) parameter.power.mc 0.93 60 0.6 0.9
#> 15 brew_temp parameter.power.mc 0.74 60 0.6 0.9
#> 16 altitude parameter.power.mc 0.73 60 0.6 0.9
#> 17 bean_sun1 parameter.power.mc 0.36 60 0.6 0.9
#> 18 bean_sun2 parameter.power.mc 0.42 60 0.6 0.9
#> 19 (Intercept) effect.power.mc 0.98 70 0.6 0.9
#> 20 brew_temp effect.power.mc 0.83 70 0.6 0.9
#> 21 altitude effect.power.mc 0.84 70 0.6 0.9
#> 22 bean_sun effect.power.mc 0.44 70 0.6 0.9
#> 23 (Intercept) parameter.power.mc 0.98 70 0.6 0.9
#> 24 brew_temp parameter.power.mc 0.83 70 0.6 0.9
#> 25 altitude parameter.power.mc 0.84 70 0.6 0.9
#> 26 bean_sun1 parameter.power.mc 0.41 70 0.6 0.9
#> 27 bean_sun2 parameter.power.mc 0.54 70 0.6 0.9
#> 28 (Intercept) effect.power.mc 1.00 80 0.6 0.9
#> 29 brew_temp effect.power.mc 0.92 80 0.6 0.9
#> 30 altitude effect.power.mc 0.86 80 0.6 0.9
#> random_seed
#> 1 123
#> 2 123
#> 3 123
#> 4 123
#> 5 123
#> 6 123
#> 7 123
#> 8 123
#> 9 123
#> 10 123
#> 11 123
#> 12 123
#> 13 123
#> 14 123
#> 15 123
#> 16 123
#> 17 123
#> 18 123
#> 19 123
#> 20 123
#> 21 123
#> 22 123
#> 23 123
#> 24 123
#> 25 123
#> 26 123
#> 27 123
#> 28 123
#> 29 123
#> 30 123
#> Power curve generation captured the following warning/error messages:
#> Function | Type | N | Message
#> Evaluation | Warn | 1 | Message: 'skpr: Partial or complete separation likely detected in the binomial Monte Carlo simulation. Increase the number of runs in the design or decrease the number of model parameters to improve power.'
if(skpr:::run_documentation()) {
#Generate power curve for a binomial model and multiple effect sizes
calculate_power_curves(trials=seq(50,150,by=10),
candidateset = cand_set,
model = ~.,
effectsize = list(c(0.5,0.9),c(0.6,0.9)),
eval_function = "eval_design_mc",
eval_args = list(nsim = 100, glmfamily = "binomial")) |>
head(30)
}
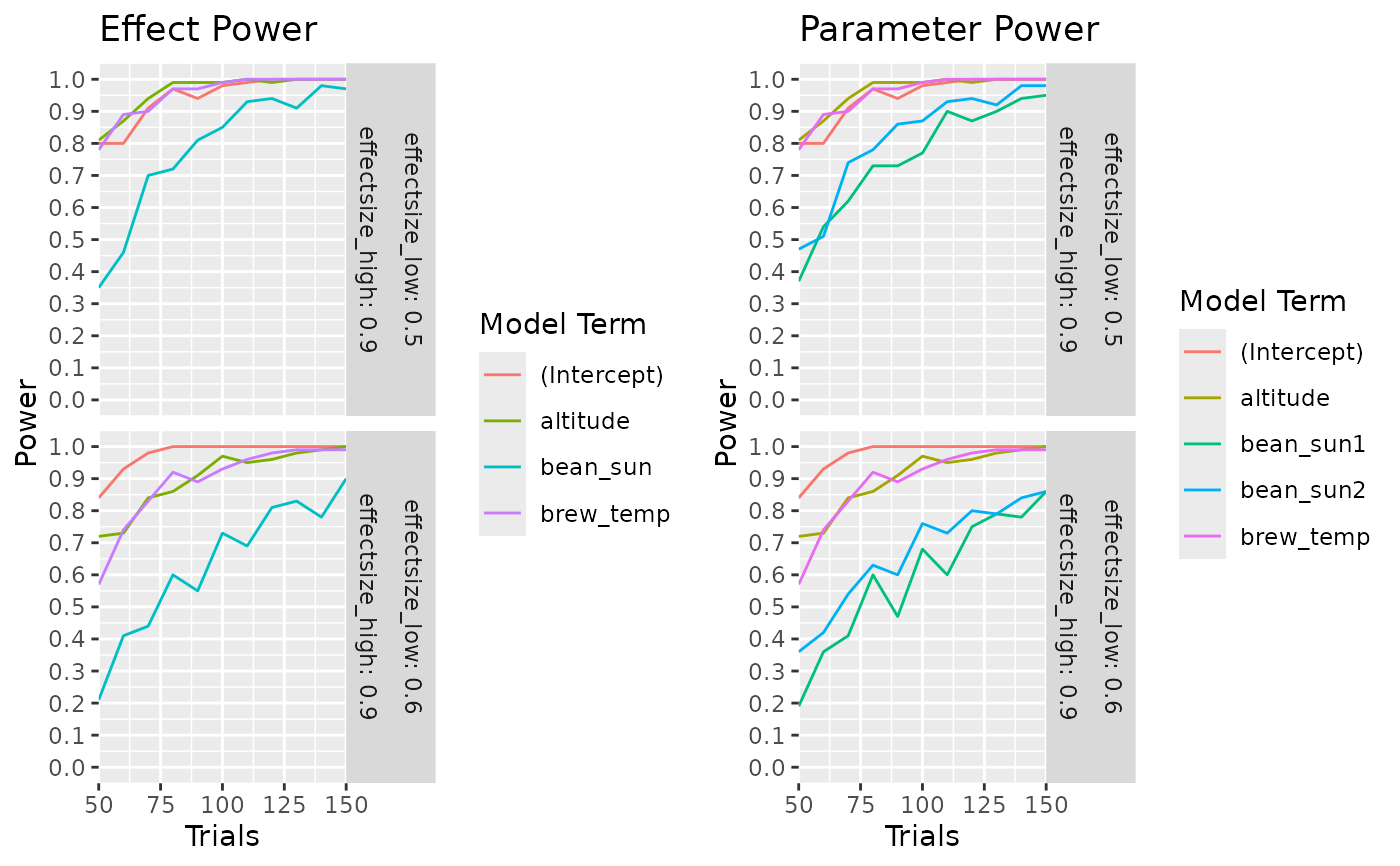 #> parameter type power trials effectsize_low effectsize_high
#> 1 (Intercept) effect.power.mc 0.80 50 0.5 0.9
#> 2 brew_temp effect.power.mc 0.78 50 0.5 0.9
#> 3 altitude effect.power.mc 0.81 50 0.5 0.9
#> 4 bean_sun effect.power.mc 0.35 50 0.5 0.9
#> 5 (Intercept) parameter.power.mc 0.80 50 0.5 0.9
#> 6 brew_temp parameter.power.mc 0.78 50 0.5 0.9
#> 7 altitude parameter.power.mc 0.81 50 0.5 0.9
#> 8 bean_sun1 parameter.power.mc 0.37 50 0.5 0.9
#> 9 bean_sun2 parameter.power.mc 0.47 50 0.5 0.9
#> 10 (Intercept) effect.power.mc 0.80 60 0.5 0.9
#> 11 brew_temp effect.power.mc 0.89 60 0.5 0.9
#> 12 altitude effect.power.mc 0.87 60 0.5 0.9
#> 13 bean_sun effect.power.mc 0.46 60 0.5 0.9
#> 14 (Intercept) parameter.power.mc 0.80 60 0.5 0.9
#> 15 brew_temp parameter.power.mc 0.89 60 0.5 0.9
#> 16 altitude parameter.power.mc 0.87 60 0.5 0.9
#> 17 bean_sun1 parameter.power.mc 0.54 60 0.5 0.9
#> 18 bean_sun2 parameter.power.mc 0.51 60 0.5 0.9
#> 19 (Intercept) effect.power.mc 0.91 70 0.5 0.9
#> 20 brew_temp effect.power.mc 0.90 70 0.5 0.9
#> 21 altitude effect.power.mc 0.94 70 0.5 0.9
#> 22 bean_sun effect.power.mc 0.70 70 0.5 0.9
#> 23 (Intercept) parameter.power.mc 0.91 70 0.5 0.9
#> 24 brew_temp parameter.power.mc 0.90 70 0.5 0.9
#> 25 altitude parameter.power.mc 0.94 70 0.5 0.9
#> 26 bean_sun1 parameter.power.mc 0.62 70 0.5 0.9
#> 27 bean_sun2 parameter.power.mc 0.74 70 0.5 0.9
#> 28 (Intercept) effect.power.mc 0.97 80 0.5 0.9
#> 29 brew_temp effect.power.mc 0.97 80 0.5 0.9
#> 30 altitude effect.power.mc 0.99 80 0.5 0.9
#> random_seed
#> 1 123
#> 2 123
#> 3 123
#> 4 123
#> 5 123
#> 6 123
#> 7 123
#> 8 123
#> 9 123
#> 10 123
#> 11 123
#> 12 123
#> 13 123
#> 14 123
#> 15 123
#> 16 123
#> 17 123
#> 18 123
#> 19 123
#> 20 123
#> 21 123
#> 22 123
#> 23 123
#> 24 123
#> 25 123
#> 26 123
#> 27 123
#> 28 123
#> 29 123
#> 30 123
#> Power curve generation captured the following warning/error messages:
#> Function | Type | N | Message
#> Evaluation | Warn | 2 | Message: 'skpr: Partial or complete separation likely detected in the binomial Monte Carlo simulation. Increase the number of runs in the design or decrease the number of model parameters to improve power.'
#> parameter type power trials effectsize_low effectsize_high
#> 1 (Intercept) effect.power.mc 0.80 50 0.5 0.9
#> 2 brew_temp effect.power.mc 0.78 50 0.5 0.9
#> 3 altitude effect.power.mc 0.81 50 0.5 0.9
#> 4 bean_sun effect.power.mc 0.35 50 0.5 0.9
#> 5 (Intercept) parameter.power.mc 0.80 50 0.5 0.9
#> 6 brew_temp parameter.power.mc 0.78 50 0.5 0.9
#> 7 altitude parameter.power.mc 0.81 50 0.5 0.9
#> 8 bean_sun1 parameter.power.mc 0.37 50 0.5 0.9
#> 9 bean_sun2 parameter.power.mc 0.47 50 0.5 0.9
#> 10 (Intercept) effect.power.mc 0.80 60 0.5 0.9
#> 11 brew_temp effect.power.mc 0.89 60 0.5 0.9
#> 12 altitude effect.power.mc 0.87 60 0.5 0.9
#> 13 bean_sun effect.power.mc 0.46 60 0.5 0.9
#> 14 (Intercept) parameter.power.mc 0.80 60 0.5 0.9
#> 15 brew_temp parameter.power.mc 0.89 60 0.5 0.9
#> 16 altitude parameter.power.mc 0.87 60 0.5 0.9
#> 17 bean_sun1 parameter.power.mc 0.54 60 0.5 0.9
#> 18 bean_sun2 parameter.power.mc 0.51 60 0.5 0.9
#> 19 (Intercept) effect.power.mc 0.91 70 0.5 0.9
#> 20 brew_temp effect.power.mc 0.90 70 0.5 0.9
#> 21 altitude effect.power.mc 0.94 70 0.5 0.9
#> 22 bean_sun effect.power.mc 0.70 70 0.5 0.9
#> 23 (Intercept) parameter.power.mc 0.91 70 0.5 0.9
#> 24 brew_temp parameter.power.mc 0.90 70 0.5 0.9
#> 25 altitude parameter.power.mc 0.94 70 0.5 0.9
#> 26 bean_sun1 parameter.power.mc 0.62 70 0.5 0.9
#> 27 bean_sun2 parameter.power.mc 0.74 70 0.5 0.9
#> 28 (Intercept) effect.power.mc 0.97 80 0.5 0.9
#> 29 brew_temp effect.power.mc 0.97 80 0.5 0.9
#> 30 altitude effect.power.mc 0.99 80 0.5 0.9
#> random_seed
#> 1 123
#> 2 123
#> 3 123
#> 4 123
#> 5 123
#> 6 123
#> 7 123
#> 8 123
#> 9 123
#> 10 123
#> 11 123
#> 12 123
#> 13 123
#> 14 123
#> 15 123
#> 16 123
#> 17 123
#> 18 123
#> 19 123
#> 20 123
#> 21 123
#> 22 123
#> 23 123
#> 24 123
#> 25 123
#> 26 123
#> 27 123
#> 28 123
#> 29 123
#> 30 123
#> Power curve generation captured the following warning/error messages:
#> Function | Type | N | Message
#> Evaluation | Warn | 2 | Message: 'skpr: Partial or complete separation likely detected in the binomial Monte Carlo simulation. Increase the number of runs in the design or decrease the number of model parameters to improve power.'